
Protein-Protein and Protein-Membrane Interactions in Cell Signaling of small GTPases, Plexin and Eph receptors
Our research focuses on protein-protein and protein-membrane interactions in cell signaling and migration in organ development, cancer, Alzheimer's and macular degeneration. For this we characterize protein interactions and seek to determine how they form the molecular basis of mechanisms in cell signaling. For these studies, we use a range of structural biology, computational as well as biophysics tools, and we also collaborate with cell biologists. The conformation, configuration as well as fluctuations of protein domains are important for signal transduction. For example, what protein structural and dynamic features give rise to binding affinity and to the specificity that selects one protein binding partner over others? Our studies will give us insight into the normal functions of the signaling proteins and how they are disrupted in diseased states. The knowledge gained will also help us in screening for small molecule agents that can be used to manipulate protein-protein interactions (and consequently signaling in cells) in a chemical biology approach. Furthermore, we develop methods that assist in the identification of interactions in protein complexes experimentally and improve their structural representation and characterization by molecular dynamics calculations. Of late our focus has expanded to include the transmembrane helices of receptors and the interaction of membrane proximal domains with the lipid bilayer. Together with the behavior of the soluble domains these interactions will explain how signals are transmitted across the cellular membrane.
Cell guidance receptors and their ligands
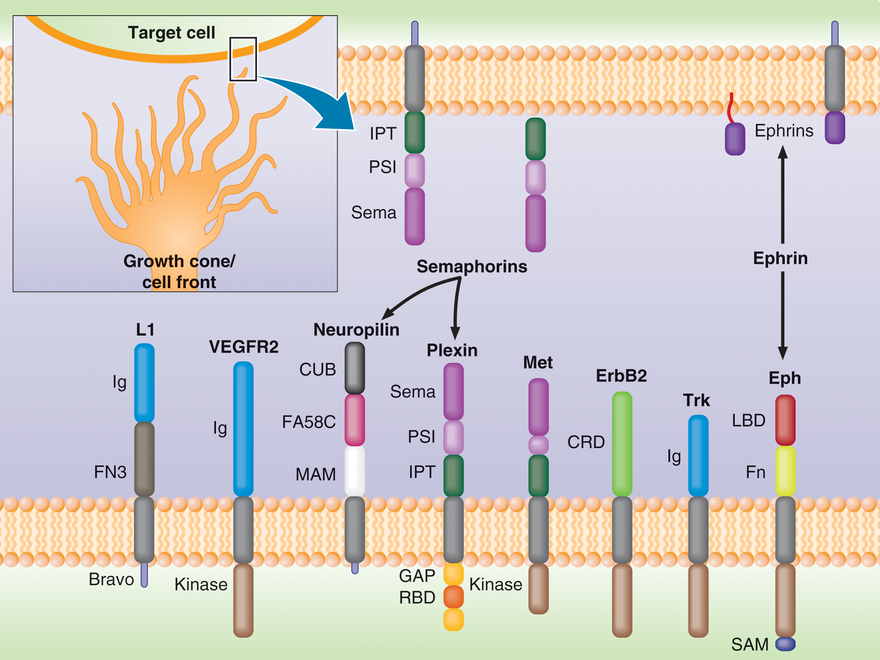
Figure Legend: Cell migration is guided by ligands that are secreted by target cells or attached to their membranes (top). The cell front of migrating cells, e.g. axonal growth cones (bottom), exhibits a number of transmembrane receptor families which bind these ligands (Semaphorins bind to Plexin and Neuroplin; Ephrin guidance cues bind to Eph receptors). Plexins also have a number of co-receptors, such as Met and ErbB2, which are involved in cancer cell migration and metastasis. We are trying to understand how the signals are transmitted across the cellular membrane and how the receptor’s intracellular domains interact with the cytoplasmic proteins that regulate cell migration.
Structure and Signaling Functions of the Plexin Transmembrane Receptor
This class of proteins receive guidance cues (such as binding of Semaphorin ligands, see Figure above) and is activated by them. The cues allow the cells to decide whether to move towards another cell, or away from it. This process of guidance, e.g. through the migration of a growth cone, is in part responsible for "wiring up" the nervous and the cardiovascular systems. The same family of receptors is also important in immunology and cancer. Plexins are unique, as they are the first example of a receptor that interacts directly with small GTPases, a family of proteins that are essential for cell migration and proliferation/survival. We have determined the structure of the Rho GTPase binding domain of several Plexins and in collaboration, also of the entire intracellular region of a Plexin. We are now characterizing how Rho GTPases influence the function of the receptor. Using the structural information we and others have acquired, we are also altering the binding affinity and specificity of plexin towards various GTPases.
Structure and Signaling Functions of the Eph-A1, -A2 and -B1 Transmembrane Receptors
Eph receptors comprise a highly abundant family of Receptor Tyrosine Kinases, also involved - similar to Plexins - in cell guidance, attachment and proliferation (see Figure, above). Although the structures of several of the cytoplasmic domains have been determined, the overall protein conformational changes that take place in signaling are not yet understood.
Role of Phosphorylation in small GTPase Signaling.
Using an in vitro protein biophysics approach we are characterizing phosphorylation patterns in regulatory and effector proteins that result from their exposure to a variety of active Ser/Thr as well as Tyr kinases. Biochemical assays put a limit on the effects that are observed on the protein’s function as a consequence of the phosphorylation. The relevant sites are then mutated and the role of the phosphorylation is verified in vivo.
| Major Research Areas |
|---|
| Angiogenesis, Biochemical Pathways, Development of Therapeutics, Neurological Disorders, Neuronal Damage, Protein Structure/Function, Protein-Protein Interactions |
| Disciplines |
| Biophysics, Cryo-Electron Microscopy, Fluorescence Spectroscopy, Hydrogen-Deuterium Exchange, Molecular Biology, Molecular Modeling and Dynamics Simulation, Nuclear Magentic Resonance , Signal Transduction, X-Ray Crystallography |
| Organ Systems |
| Cardiovascular System, Nervous System, Renal System |
| Diseases |
| Alzheimer's Disease, Cancer, Cardiovascular Regeneration, Neurodegenerative Diseases, Parkinson's Disease |
| Major Techniques |
| 31P Nuclear Magnetic Resonance Spectroscopy, Cellular and Molecular Biology, Cryo-Electron Microscopy, EPR, Gel Electrophoresis/Western Blots, Mass Spectrometry, Molecular Dynamics, NMR, Protein Crystallization, Protein Expression, Protein-Protein Interactions (Biocore), Spectrophotometry |